Getting started#
Requirements#
A recent version of Matlab
Downloading and Installing#
Download the current version of the entire Pulmonary Toolkit and place in a folder on your Matlab path.
If you have a C++ compiler, you should ensure Matlab is set up to use it by calling
mex -setup
in the Matlab command window, and follow the instructions to set up Matlab’s mex compiler.
The C++ compiler is optional, but segmentation features of the Pulmonary Toolkit (alpha version) will not function without it.
Note: The installer script sets up Matlab’s C++ mex compiler and uses this to compile certain functions used by the Toolkit. For more information about mex, typedoc mexat the Matlab command window. For Windows and Mac you may need to install a C++ compiler before running the install script. XCode (for Mac) or Visual Studio (for Windows) will install compilers on your system.
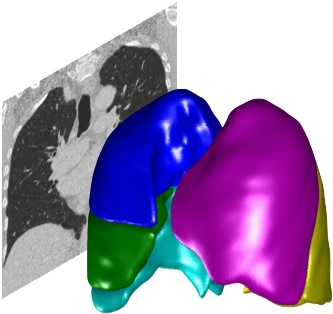
Data#
The toolkit is designed for use with high-resolution CT and MRI clinical lung images in 3D. Other types of image may work but are not officially supported.
Data can be imported in a variety of formats. You should import from original DICOM files where available, so that the toolkit can make use of its metadata.
PTKViewer##
PTKViewer is a standalone application for visualising 3D or 2D data slice-by-slice. See the page PTKViewer for more information
Running the Pulmonary Toolkit user interface#
To run the PTK gui, run
ptk
The use the Import button to import data