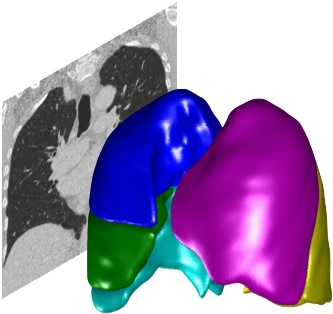
Supported data and imaging formats#
PTK is primarily designed to work with 3D CT and MRI images covering the whole lung.
You may be able to load and visualise other data into PTK, but you may find the segmentation and other Plugins do not work.
Data not covering the whole lung#
The PTK algorithms work best when the 3D volume covers the entire lung. There are adjustment mechanisms in place which allow for lung boundaries to extend outside the image in some cases, although this could result in inaccurate measurements such as for lung volume.
Non-lung data#
If you want to work on non-lung images, you might consider using the provided MIV (Medical Imaging Viewer), which is similar to PTK but is not lung-specific.
Supported data formats#
DICOM#
PTK uses DicomMat to group DICOM images into Datasets. This allows PTK to separate out scout or report images from the core image files, and to separate non-contiguous datasets.
The DICOM Series Instance UID tag is used to uniquely identify the dataset, so this must be unique for each series (as required by the DICOM standard). If DICOM data are imported with a series instance UID that matches an existing dataset in PTK, the behaviour is undefined.
MetaIO#
MetaIO files are usually a single .mha file, or a pair of files eg. .mhd & .raw.
Other formats#
Limited support is provided for other formats. Some of these use the Read Medical Data 3D library by Dirk-Jan Kroon. The use of other formats has not been fully tested in PTK so you may find that not all functionality works.